2×2 repeated measures ANOVAs are common in my work. For that, I now created a flexible function in R. The function outputs assumption checks (outliers and normality), interaction and main effect results, pairwise comparisons, and produces a result plot with within-subject error bars (SD, SE or 95% CI) and significance stars added to the plot.
The function rm_2by2_anova.R and its accompanying notebook is available here
https://github.com/dcdace/R_functions
Below is the HTML version of the notebook. In the notebook, I test the function on three different datasets and also provide a step-by-step walk through the function elements.
Having spent many hours on this I hope the function will be useful and time-saving also to others.
Please let me know if you find any statistics or other errors!
Table of Contents
- 1 Example 1: Using rm_2by2_anova function
- 2 Example 2: Step-by-step example
- 3 Example 3: Test on another dataset
- 4 Example 4: Test on another dataset
- 5 Control what to display
2x2 Repeated Measures ANOVA¶
# Dace Apšvalka, 2020, www.dcdace.net
# ----------------------------------------------------------------------
# LIBRARIES
# ----------------------------------------------------------------------
shhh <- suppressPackageStartupMessages # It's a library, so shhh!
shhh(suppressWarnings(library(psycho))) # to get the Emotion dataset for the example
Example 1: Using rm_2by2_anova function¶
Load the function¶
source("https://raw.githubusercontent.com/dcdace/R_functions/main/RM-2by2-ANOVA/rm_2by2_anova.R")
Load example data¶
# ----------------------------------------------------------------------
# Get the data
# ----------------------------------------------------------------------
# As an example I use Emotion dataset from psycho package, https://github.com/neuropsychology/psycho.R
# Get the dataset
dataset <- psycho::emotion
# See how it looks
head(dataset)
| Participant_ID | Participant_Age | Participant_Sex | Item_Category | Item_Name | Trial_Order | Emotion_Condition | Subjective_Arousal | Subjective_Valence | Autobiographical_Link | Recall | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| <fct> | <dbl> | <fct> | <fct> | <fct> | <dbl> | <fct> | <dbl> | <dbl> | <dbl> | <lgl> | |
| 1 | 1S | 18.38467 | Female | People | People_158_h | 1 | Neutral | 12.239583 | 0.5208333 | 63.281250 | TRUE |
| 2 | 1S | 18.38467 | Female | Faces | Faces_045_h | 2 | Neutral | 16.406250 | 4.1666667 | 22.135417 | FALSE |
| 3 | 1S | 18.38467 | Female | People | People_138_h | 3 | Neutral | 25.520833 | 25.5208333 | 55.989583 | TRUE |
| 4 | 1S | 18.38467 | Female | People | People_148_h | 4 | Neutral | 0.000000 | 0.0000000 | 44.791667 | FALSE |
| 5 | 1S | 18.38467 | Female | Faces | Faces_315_h | 5 | Neutral | 25.781250 | 45.8333333 | 30.208333 | FALSE |
| 6 | 1S | 18.38467 | Female | Faces | Faces_224_h | 6 | Neutral | 2.604167 | 0.0000000 | 8.072917 | FALSE |
Input data for the function¶
# ----------------------------------------------------------------------
# Specify items and parameters
# ----------------------------------------------------------------------
# From the dataset, specify subject ID, Dependent Variable, and 2 within-subject Factors
columns <- list(
sID = "Participant_ID", # subject ID
DV = "Subjective_Valence", # dependent variable
Fc1 = "Item_Category", # within-subj factor 1
Fc2 = "Emotion_Condition" # within-subj factor 2
)
# Define plot label names, tilte, colors and what type of error bars to show on the plot
param <- list(
y.label = "Subjective Valence",
Fc1.label = "Item",
Fc2.label = "Emotion",
cat.color = c('#DF4A56', '#5284a8'),
errorbar = "ci" # can be either sd, se, or ci
)
# the plot title
param$title <- sprintf("%s x %s interaction", param$Fc2.label, param$Fc1.label)
Run the function¶
The function will output several result summaries. And it will also return a list of result variables including assumption plot (boxplots & QQ plot) and the interaction plot.
results <- rm_2by2_anova(dataset, columns, param)
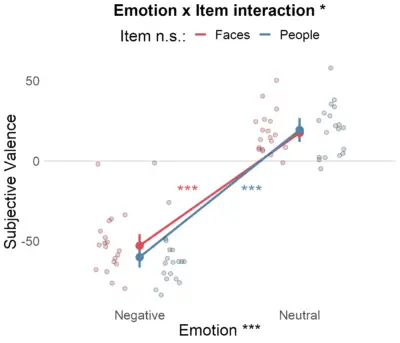
================================================================ CHECK ASSUMPTIONS ================================================================ Outliers # A tibble: 4 x 6 Fc1 Fc2 sID meanDV is.outlier is.extreme <fct> <fct> <fct> <dbl> <lgl> <lgl> 1 Faces Negative 17S -1.91 TRUE FALSE 2 Faces Neutral 9S 50.5 TRUE FALSE 3 People Negative 17S -1.17 TRUE TRUE 4 People Negative 9S -25.8 TRUE FALSE Data has extreme outliers! ----------------------------------------------- Normality # A tibble: 4 x 5 Fc1 Fc2 variable statistic p <fct> <fct> <chr> <dbl> <dbl> 1 Faces Negative meanDV 0.928 0.159 2 Faces Neutral meanDV 0.926 0.145 3 People Negative meanDV 0.833 0.00364 4 People Neutral meanDV 0.947 0.351 Not all levels are normaly distributed! ================================================================ SUMMARY DESCRIPTIVES ================================================================ Fc1 Fc2 N meanDV sd se ci 1 Faces Negative 19 -52.75116 14.77499 3.389615 7.121317 2 Faces Neutral 19 17.62804 11.73257 2.691635 5.654916 3 People Negative 19 -59.92947 13.65515 3.132706 6.581571 4 People Neutral 19 19.45238 15.63017 3.585806 7.533500 ================================================================ 2x2 INTERACTION RESULTS ================================================================ Error: sID Df Sum Sq Mean Sq F value Pr(>F) Residuals 18 9623 534.6 Error: sID:Fc1 Df Sum Sq Mean Sq F value Pr(>F) Fc1 1 136.2 136.16 1.499 0.237 Residuals 18 1634.6 90.81 Error: sID:Fc2 Df Sum Sq Mean Sq F value Pr(>F) Fc2 1 106535 106535 245.9 6.11e-12 *** Residuals 18 7798 433 --- Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1 Error: sID:Fc1:Fc2 Df Sum Sq Mean Sq F value Pr(>F) Fc1:Fc2 1 385 385 5.831 0.0266 * Residuals 18 1188 66 --- Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1 Significant Emotion x Item interaction: F(1,18) = 5.83, p = 0.0266076 ================================================================ Main Effects ================================================================ Main effect of Item: F(1,18) = 1.50, p = 0.2365520 ----------------------------------------------- Main effect of Emotion: F(1,18) = 245.92, p = 0.0000000 ================================================================ Post-hoc ================================================================ Pairwise comparisons grouped by Factor 1 # A tibble: 2 x 11 Fc1 .y. group1 group2 n1 n2 statistic df p p.adj * <fct> <chr> <chr> <chr> <int> <int> <dbl> <dbl> <dbl> <dbl> 1 Faces meanDV Negative Neutral 19 19 -14.6 18 2.09e-11 2.09e-11 2 People meanDV Negative Neutral 19 19 -14.7 18 1.84e-11 1.84e-11 # ... with 1 more variable: p.adj.signif <chr> ----------------------------------------------- Pairwise comparisons grouped by Factor 2 # A tibble: 2 x 11 Fc2 .y. group1 group2 n1 n2 statistic df p p.adj p.adj.signif * <fct> <chr> <chr> <chr> <int> <int> <dbl> <dbl> <dbl> <dbl> <chr> 1 Nega~ mean~ Faces People 19 19 2.37 18 0.029 0.029 * 2 Neut~ mean~ Faces People 19 19 -0.673 18 0.51 0.51 ns
Assumption check plots¶
options(repr.plot.width = 10, repr.plot.height = 5, repr.plot.res = 200) # change plot size
results$plot.assumption.checks
Results plot¶
Error bars in the plot are within-subject 95% CIs.
Basic plot¶
options(repr.plot.width = 7, repr.plot.height = 5, repr.plot.res = 200) # change plot size
results$plot.anova
Add some extra stuff to the plot¶
#options(repr.plot.width = 7, repr.plot.height = 6, repr.plot.res = 200) # change plot size
# Adding post-hoc comparison stars
# Prepare the values
# post-hoc pairwise comparisons for each Fc1 level between the Fc2 levels
p.Fc1.L1 <-
ifelse(results$pwc1$p[1] < 0.05, stars.pval(results$pwc1$p[1]), "n.s.")
p.Fc1.L2 <-
ifelse(results$pwc1$p[2] < 0.05, stars.pval(results$pwc1$p[2]), "n.s.")
# add horizontal line BEHIND other elements
results$plot.anova$layers <- c(geom_hline(aes(yintercept = 0), size = 0.1), # the new layer
results$plot.anova$layers) # original layers
# add other things
results$plot.anova +
# Add post-hoc pairwise comparisons
# might need to adjust the x and y positions
annotate(
"text",
x = 1.30,
y = mean(results$dataSummary$meanDV),
label = p.Fc1.L1,
color = param$cat.color[1],
size = 8
) +
annotate(
"text",
x = 1.7,
y = mean(results$dataSummary$meanDV),
label = p.Fc1.L2,
color = param$cat.color[2],
size = 8
) +
# add individual datapoints
geom_point(
colour = "black",
alpha = .3,
aes(fill = Fc1),
size = 2,
stroke = 0.5,
shape = 21,
position = position_jitterdodge(),
show.legend = FALSE
)
Example 2: Step-by-step example¶
Here the above function elements are executed ste-by-step (the function itself is not required).
Specify parameters¶
# ----------------------------------------------------------------------
# Specify items and parameters
# ----------------------------------------------------------------------
# From the dataset, specify subject ID, Dependent Variable, and 2 within-subject Factors
columns <- list(
sID = "Participant_ID", # subject ID
DV = "Subjective_Valence", # dependent variable
Fc1 = "Item_Category", # within-subj factor 1
Fc2 = "Emotion_Condition" # within-subj factor 2
)
# Define plot label names, tilte, colors and what type of error bars to show on the plot
param <- list(
y.label = "Subjective Valence",
Fc1.label = "Item",
Fc2.label = "Emotion",
cat.color = c('#DF4A56', '#5284a8'),
errorbar = "ci" # can be either sd, se, or ci
)
# the plot title
param$title <- sprintf("%s x %s interaction", param$Fc2.label, param$Fc1.label)
Prepare the dataset¶
# ----------------------------------------------------------------------
# Prepare the working dataset
# ----------------------------------------------------------------------
# Subset the dataset to only the columns of interest
# Because dplyr::select function clashes with MASS::select need to specify to use dplyr:: select
data_subset <- dataset %>% dplyr::select(all_of(unlist(columns)))
# Average same Factor/Level values for each subject if there are several.
# Will use this data frame df for the rest of the results and plots
df <- ddply(data_subset, .(sID, Fc1, Fc2),
summarise, meanDV = mean(DV, na.rm = TRUE))
# sID, Fc1 and Fc2 must be factors (in case they are not already)
df$sID <- as.factor(df$sID)
df$Fc1 <- as.factor(df$Fc1)
df$Fc2 <- as.factor(df$Fc2)
# See how it looks
head(df)
| sID | Fc1 | Fc2 | meanDV | |
|---|---|---|---|---|
| <fct> | <fct> | <fct> | <dbl> | |
| 1 | 10S | Faces | Negative | -53.472222 |
| 2 | 10S | Faces | Neutral | 32.508681 |
| 3 | 10S | People | Negative | -62.673611 |
| 4 | 10S | People | Neutral | 20.833333 |
| 5 | 11S | Faces | Negative | -76.189605 |
| 6 | 11S | Faces | Neutral | 8.388238 |
Check assumptions¶
Outliers¶
# ----------------------------------------------------------------------
# Check for outliers
# ----------------------------------------------------------------------
# Identify if there are extreme outliers
check.outliers <- df %>%
group_by(Fc1, Fc2) %>%
identify_outliers(meanDV)
# Display
check.outliers
# Outlier result text
outlier.result.txt <- ifelse(any(check.outliers$is.extreme == T),
"Data has extreme outliers!",
"There are no extreme outliers.")
cat("================================================\n\n")
cat(outlier.result.txt)
cat("\n\n================================================")
# Boxplot title color. It will be red if there are ouliers!
outlier.title.color <- ifelse(any(check.outliers$is.extreme == T),
"red",
"black")
# Create a boxplot
plot.box <- ggboxplot(
df, x = "Fc2", y = "meanDV",
color = "Fc1",
title = outlier.result.txt,
ylab = param$y.label,
xlab = param$Fc2.label
) +
guides(color = guide_legend(param$Fc1.label)) +
scale_color_manual(values = param$cat.color) +
theme(plot.title = element_text(hjust = 0.5, color = outlier.title.color))
| Fc1 | Fc2 | sID | meanDV | is.outlier | is.extreme |
|---|---|---|---|---|---|
| <fct> | <fct> | <fct> | <dbl> | <lgl> | <lgl> |
| Faces | Negative | 17S | -1.909722 | TRUE | FALSE |
| Faces | Neutral | 9S | 50.469741 | TRUE | FALSE |
| People | Negative | 17S | -1.171875 | TRUE | TRUE |
| People | Negative | 9S | -25.817472 | TRUE | FALSE |
================================================ Data has extreme outliers! ================================================
Normality¶
# ----------------------------------------------------------------------
# Check normality assumption
# ----------------------------------------------------------------------
# Shapiro-Wilk’s test (should be > .05)
check.normality <- df %>%
group_by(Fc1, Fc2) %>%
shapiro_test(meanDV)
# Display
check.normality
# Normality result text
normality.result.txt <- ifelse(any(check.normality$p < 0.05),
"Not all levels are normaly distributed!",
"All levels are normally distributed.")
cat("================================================\n\n")
cat(normality.result.txt)
cat("\n\n================================================")
# QQ plot title color. It will be red if not all levels meet normality!
normality.title.color <- ifelse(any(check.normality$p < 0.05),
"red",
"black")
# Create Quantile-Quantile plots
plot.qq <- ggqqplot(df, "meanDV", ggtheme = theme_minimal(), title = normality.result.txt) +
facet_grid(Fc1 ~ Fc2, labeller = "label_both") +
theme(plot.title = element_text(hjust = 0.5, color = normality.title.color))
| Fc1 | Fc2 | variable | statistic | p |
|---|---|---|---|---|
| <fct> | <fct> | <chr> | <dbl> | <dbl> |
| Faces | Negative | meanDV | 0.9279360 | 0.158654565 |
| Faces | Neutral | meanDV | 0.9258916 | 0.145440020 |
| People | Negative | meanDV | 0.8333678 | 0.003640378 |
| People | Neutral | meanDV | 0.9469856 | 0.350725108 |
================================================ Not all levels are normaly distributed! ================================================
Boxplot and QQ plots¶
# ----------------------------------------------------------------------
# PLOT Boxplot and QQ
# ----------------------------------------------------------------------
options(repr.plot.width = 10, repr.plot.height = 5, repr.plot.res = 200) # change plot size
# Add result texts as titles for each plot
plot_grid(plot.box, plot.qq, nrow = 1, rel_widths = c(1/4,3/4))
Descriptive summary¶
# ----------------------------------------------------------------------
# Within-Subject descriptive summary
# ----------------------------------------------------------------------
# Get the within-subject summary. Will need this to plot within-subject error bars.
# Using summarySEwithin function from the Rmisc package
dataSummary <- summarySEwithin(
df,
measurevar = "meanDV",
withinvars = c("Fc1", "Fc2"),
idvar = "sID",
na.rm = TRUE,
conf.interval = 0.95
)
# See how it looks
dataSummary
| Fc1 | Fc2 | N | meanDV | sd | se | ci |
|---|---|---|---|---|---|---|
| <fct> | <fct> | <dbl> | <dbl> | <dbl> | <dbl> | <dbl> |
| Faces | Negative | 19 | -52.75116 | 14.77499 | 3.389615 | 7.121317 |
| Faces | Neutral | 19 | 17.62804 | 11.73257 | 2.691635 | 5.654916 |
| People | Negative | 19 | -59.92947 | 13.65515 | 3.132706 | 6.581571 |
| People | Neutral | 19 | 19.45238 | 15.63017 | 3.585806 | 7.533500 |
2 x 2 ANOVA results¶
2-way interaction¶
# ----------------------------------------------------------------------
# Factor 1 X Factor 2 interaction, ANOVA
# ----------------------------------------------------------------------
# Within-subject repeated measures ANOVA
results$res.anova <-
aov(meanDV ~ Fc1 * Fc2 + Error(sID / (Fc1 * Fc2)),
data = df)
#Summary results
summary.anova <- summary(results$res.anova)
# save p-val, will need it to put on the plot
res.anova.pval <-
summary.anova[["Error: sID:Fc1:Fc2"]][[1]][["Pr(>F)"]][1]
# Display result string
sign.anova <-
ifelse(res.anova.pval < 0.05, "Significant", "No significant")
results$res.txt.anova <- sprintf(
"\n%s %s: F(%d,%d) = %.2f, p = %.7f\n",
sign.anova,
param$title,
summary.anova[["Error: sID:Fc1:Fc2"]][[1]][["Df"]][1],
summary.anova[["Error: sID:Fc1:Fc2"]][[1]][["Df"]][2],
summary.anova[["Error: sID:Fc1:Fc2"]][[1]][["F value"]][1],
res.anova.pval
)
cat("\n================================================\n\n")
cat(results$res.txt.anova)
cat("\n\n================================================")
================================================ Significant Emotion x Item interaction: F(1,18) = 5.83, p = 0.0266076 ================================================
Main Effects of Factor 1¶
# Main Effect of Factor 1
# save p-val, will need it to put on the plot
res.Fc1.pval <-
summary.anova[["Error: sID:Fc1"]][[1]][["Pr(>F)"]][1]
# Display result string
sign.Fc1 <-
ifelse(res.Fc1.pval < 0.05, "Significant", "No significant")
results$res.txt.Fc1 <- sprintf(
"Main effect of %s: F(%d,%d) = %.2f, p = %.7f\n",
param$Fc1.label,
summary.anova[["Error: sID:Fc1"]][[1]][["Df"]][1],
summary.anova[["Error: sID:Fc1"]][[1]][["Df"]][2],
summary.anova[["Error: sID:Fc1"]][[1]][["F value"]][1],
summary.anova[["Error: sID:Fc1"]][[1]][["Pr(>F)"]][1]
)
# display the result sentence
cat(results$res.txt.Fc1)
Main effect of Item: F(1,18) = 1.50, p = 0.2365520
Main Effects of Factor 2¶
# Main Effect of Factor 2
res.Fc2.pval <-
summary.anova[["Error: sID:Fc2"]][[1]][["Pr(>F)"]][1]
# Display result string
sign.Fc2 <-
ifelse(res.Fc2.pval < 0.05, "Significant", "No significant")
results$res.txt.Fc2 <- sprintf(
"Main effect of %s: F(%d,%d) = %.2f, p = %.7f\n",
param$Fc2.label,
summary.anova[["Error: sID:Fc2"]][[1]][["Df"]][1],
summary.anova[["Error: sID:Fc2"]][[1]][["Df"]][2],
summary.anova[["Error: sID:Fc2"]][[1]][["F value"]][1],
summary.anova[["Error: sID:Fc2"]][[1]][["Pr(>F)"]][1]
)
# display the result sentence
cat(results$res.txt.Fc2)
Main effect of Emotion: F(1,18) = 245.92, p = 0.0000000
Post-hoc tests¶
# Pairwise comparisons grouped by Factor 1
pwc1 <- df %>%
group_by(Fc1) %>%
pairwise_t_test(
meanDV ~ Fc2, paired = TRUE
)
pwc1
| Fc1 | .y. | group1 | group2 | n1 | n2 | statistic | df | p | p.adj | p.adj.signif | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| <fct> | <chr> | <chr> | <chr> | <int> | <int> | <dbl> | <dbl> | <dbl> | <dbl> | <chr> | |
| 1 | Faces | meanDV | Negative | Neutral | 19 | 19 | -14.57339 | 18 | 2.09e-11 | 2.09e-11 | **** |
| 2 | People | meanDV | Negative | Neutral | 19 | 19 | -14.68316 | 18 | 1.84e-11 | 1.84e-11 | **** |
# Pairwise comparisons grouped by Factor 2
pwc2 <- df %>%
group_by(Fc2) %>%
pairwise_t_test(
meanDV ~ Fc1, paired = TRUE
)
pwc2
| Fc2 | .y. | group1 | group2 | n1 | n2 | statistic | df | p | p.adj | p.adj.signif | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| <fct> | <chr> | <chr> | <chr> | <int> | <int> | <dbl> | <dbl> | <dbl> | <dbl> | <chr> | |
| 1 | Negative | meanDV | Faces | People | 19 | 19 | 2.3724765 | 18 | 0.029 | 0.029 | * |
| 2 | Neutral | meanDV | Faces | People | 19 | 19 | -0.6727361 | 18 | 0.510 | 0.510 | ns |
Plot the results¶
Preparing titles and annotations¶
# ----------------------------------------------------------------------
# P-values for displaying significance stars
# ----------------------------------------------------------------------
# 2x2 interaction result
p.anova <-
ifelse(res.anova.pval < 0.05, stars.pval(res.anova.pval), "n.s.")
# Main effects
p.mainFc1 <-
ifelse(results$one.way1$p < 0.05,
stars.pval(results$one.way1$p),
"n.s.")
p.mainFc2 <-
ifelse(results$one.way2$p < 0.05,
stars.pval(results$one.way2$p),
"n.s.")
# plot main title and factor labels (with added significance stars)
plot.title <- sprintf("%s %s", param$title, p.anova)
Fc1.title <- sprintf("%s %s:", param$Fc1.label, p.mainFc1)
Fc2.title <- sprintf("%s %s", param$Fc2.label, p.mainFc2)
The plotting function¶
# ----------------------------------------------------------------------
# Plot the interaction results
# ----------------------------------------------------------------------
plot.anova <-
ggplot(data = df,
aes(
x = Fc2,
y = meanDV,
group = Fc1,
color = Fc1
)) +
# connect the level means with a line
stat_summary(fun = mean, geom = "line", size = 1) +
# add mean point and error bars (taken from dataSummary created above)
geom_pointrange(
data = dataSummary,
aes(
y = meanDV,
ymin = meanDV - .data[[param$errorbar]],
ymax = meanDV + .data[[param$errorbar]],
color = Fc1
),
size = 1,
stroke = 0.5,
show.legend = FALSE
) +
# change the default colors to the defined ones
scale_color_manual(values = param$cat.color) +
scale_fill_manual(values = param$cat.color) +
# add labels and the title
guides(color = guide_legend(Fc1.title)) +
labs(x = Fc2.title, y = param$y.label) +
ggtitle(plot.title) +
# make it nicer
theme_minimal() +
theme(
text = element_text(size = 16),
plot.title = element_text(hjust = 0.5,
size = 16,
face = "bold"),
panel.grid.major = element_blank(),
panel.grid.minor = element_blank(),
legend.position = "top"
)
Add additional items to the plot and display the plot¶
options(repr.plot.width = 7, repr.plot.height = 6) # change plot size
eb <- "sd"
# ----------------------------------------------------------------------
# Add extra stuff to the plot
# ----------------------------------------------------------------------
plot.anova +
# add post-hoc pairwise comparisons
# might need to adjust the x and y positions
annotate(
"text", x = 1.30, y = mean(dataSummary$meanDV),
label = p.Fc1.L1, color = param$cat.color[1], size = 8
) +
annotate(
"text", x = 1.7, y = mean(dataSummary$meanDV),
label = p.Fc1.L2, color = param$cat.color[2], size = 8
) +
# add individual datapoints
geom_point(
colour = "black", alpha = .3,
aes(fill = Fc1),
size = 2, stroke = 0.5, shape = 21,
position = position_jitterdodge(),
show.legend = FALSE
) +
# add horizontal line at 0
geom_hline(aes(yintercept = 0), size = 0.1)
Example 3: Test on another dataset¶
Here I am testing the rm_2by2_anova function on another dataset. The dataset is from https://ademos.people.uic.edu/Chapter21.html. The background of the dataset is as follows:
This fake data is a sample of 30 adults. All subjects were shown a set of words. Afterwards, for some of the words they were given a multiple choice test, and the rest of the words they simply restudied. Immediately afterwards their memory was tested for all words to determine if there were memory differences depending on whether the word was tested or restudied (immediate final test). Two days later, subjects came back and were given the same test on all words again (delayed final test). The dependent variable (DV) given in the file are a proportion of correctly remembered items for the immediate and delayed test for both conditions. Therefore, if we want to know if there are memory difference based on time delay and whether the word was tested or restudied, we need to conduct a within-subjects ANOVA.
Load the data¶
# https://ademos.people.uic.edu/Chapter21.html
# load the data
study_dataset <- read.csv("https://ademos.people.uic.edu/Chapter_21_Within_Data.csv")
# Dataset content:
#Subject = Subject ID #
#Within_Cond = Study Method (test or restudy)
#Within_Time = Immediate or Delayed
#DV = Memory Performance
# Change the column names and levels to reflect the data content
study_dataset$Within_Cond <- factor(study_dataset$Within_Cond,
levels = c(1,2),
labels = c("Test", "Restudy"))
study_dataset$Within_Time <-factor(study_dataset$Within_Time,
levels = c(1,2),
labels = c("Immediate", "Delayed"))
colnames(study_dataset)[2] <- "Study_Method"
colnames(study_dataset)[3] <- "Test_Time"
colnames(study_dataset)[4] <- "Memory_Performance"
head(study_dataset)
| Subject | Study_Method | Test_Time | Memory_Performance | |
|---|---|---|---|---|
| <int> | <fct> | <fct> | <dbl> | |
| 1 | 1 | Test | Immediate | 96 |
| 2 | 2 | Test | Immediate | 81 |
| 3 | 3 | Test | Immediate | 73 |
| 4 | 4 | Test | Immediate | 90 |
| 5 | 5 | Test | Immediate | 72 |
| 6 | 6 | Test | Immediate | 83 |
Specify parameters¶
# ----------------------------------------------------------------------
# Specify items and parameters
# ----------------------------------------------------------------------
# From the dataset, specify subject ID, Dependent Variable, and 2 within-subject Factors
columns <- list(
sID = "Subject",
DV = "Memory_Performance",
Fc1 = "Study_Method",
Fc2 = "Test_Time"
)
# Define plot label names, tilte, colors and what type of error bars to show on the plot
param <- list(
y.label = "Memory Performance",
Fc1.label = "Study Method",
Fc2.label = "Test Time",
cat.color = c('darkblue', 'lightgreen'),
errorbar = "ci" # can be either sd, se, or ci
)
# the plot title
param$title <- sprintf("%s x %s interaction", param$Fc2.label, param$Fc1.label)
Run the function¶
results <- rm_2by2_anova(study_dataset, columns, param)
================================================================ CHECK ASSUMPTIONS ================================================================ Outliers [1] Fc1 Fc2 sID meanDV is.outlier is.extreme <0 rows> (or 0-length row.names) There are no extreme outliers. ----------------------------------------------- Normality # A tibble: 4 x 5 Fc1 Fc2 variable statistic p <fct> <fct> <chr> <dbl> <dbl> 1 Test Immediate meanDV 0.922 0.0301 2 Test Delayed meanDV 0.934 0.0645 3 Restudy Immediate meanDV 0.958 0.280 4 Restudy Delayed meanDV 0.871 0.00176 Not all levels are normaly distributed! ================================================================ SUMMARY DESCRIPTIVES ================================================================ Fc1 Fc2 N meanDV sd se ci 1 Test Immediate 30 81.26667 9.475361 1.729956 3.538158 2 Test Delayed 30 83.80000 9.232216 1.685564 3.447366 3 Restudy Immediate 30 83.40000 9.504108 1.735205 3.548892 4 Restudy Delayed 30 68.30000 12.424643 2.268419 4.639438 ================================================================ 2x2 INTERACTION RESULTS ================================================================ Error: sID Df Sum Sq Mean Sq F value Pr(>F) Residuals 29 2497 86.12 Error: sID:Fc1 Df Sum Sq Mean Sq F value Pr(>F) Fc1 1 1340 1340.0 12.24 0.00153 ** Residuals 29 3176 109.5 --- Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1 Error: sID:Fc2 Df Sum Sq Mean Sq F value Pr(>F) Fc2 1 1184 1184.4 13.21 0.00107 ** Residuals 29 2599 89.6 --- Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1 Error: sID:Fc1:Fc2 Df Sum Sq Mean Sq F value Pr(>F) Fc1:Fc2 1 2332 2332.0 20.16 0.000104 *** Residuals 29 3354 115.6 --- Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1 Significant Test Time x Study Method interaction: F(1,29) = 20.17, p = 0.0001042 ================================================================ Main Effects ================================================================ Main effect of Study Method: F(1,29) = 12.24, p = 0.0015321 ----------------------------------------------- Main effect of Test Time: F(1,29) = 13.21, p = 0.0010667 ================================================================ Post-hoc ================================================================ Pairwise comparisons grouped by Factor 1 # A tibble: 2 x 11 Fc1 .y. group1 group2 n1 n2 statistic df p p.adj * <fct> <chr> <chr> <chr> <int> <int> <dbl> <dbl> <dbl> <dbl> 1 Test meanDV Immediate Delayed 30 30 -1.12 29 0.271 2.71e-1 2 Restudy meanDV Immediate Delayed 30 30 5.15 29 0.0000165 1.65e-5 # ... with 1 more variable: p.adj.signif <chr> ----------------------------------------------- Pairwise comparisons grouped by Factor 2 # A tibble: 2 x 11 Fc2 .y. group1 group2 n1 n2 statistic df p p.adj * <fct> <chr> <chr> <chr> <int> <int> <dbl> <dbl> <dbl> <dbl> 1 Immediate meanDV Test Restudy 30 30 -0.870 29 0.392 3.92e-1 2 Delayed meanDV Test Restudy 30 30 5.17 29 0.0000159 1.59e-5 # ... with 1 more variable: p.adj.signif <chr>
Assumption check plots¶
options(repr.plot.width = 10, repr.plot.height = 5, repr.plot.res = 200) # change plot size
results$plot.assumption.checks
Results plot (with extras)¶
options(repr.plot.width = 7, repr.plot.height = 6, repr.plot.res = 200) # change plot size
# Adding post-hoc comparison stars
# Prepare the values
# post-hoc pairwise comparisons for each Fc1 level between the Fc2 levels
p.Fc1.L1 <-
ifelse(results$pwc1$p[1] < 0.05, stars.pval(results$pwc1$p[1]), "n.s.")
p.Fc1.L2 <-
ifelse(results$pwc1$p[2] < 0.05, stars.pval(results$pwc1$p[2]), "n.s.")
results$plot.anova +
# Add post-hoc pairwise comparisons
# might need to adjust the x and y positions
annotate(
"text",
x = 1.5,
y = 86,
label = p.Fc1.L1,
color = param$cat.color[1],
size = 8
) +
annotate(
"text",
x = 1.5,
y = 72,
label = p.Fc1.L2,
color = param$cat.color[2],
size = 8
) +
# add individual datapoints
geom_point(
colour = "black",
alpha = .3,
aes(fill = Fc1),
size = 2,
stroke = 0.5,
shape = 21,
position = position_jitterdodge(),
show.legend = FALSE
)
Example 4: Test on another dataset¶
Here I test the function on kepress learning by observation dataset. This is my dataset published in https://doi.org/10.1523/JNEUROSCI.1597-18.2018
Load the data¶
dataOL <-
read.csv("https://raw.githubusercontent.com/dcdace/E2fMRI_MVPA_PPI/master/Data/BehaviouralResults_longformat.csv")
# Change the Type levels to better reflect the data content
dataOL$Type <- factor(dataOL$Type,
levels = c("TR", "UN"),
labels = c("Trained", "Untrained"))
head(dataOL)
| sID | gender | age | righthanded | Time | Type | ET | Err | |
|---|---|---|---|---|---|---|---|---|
| <int> | <chr> | <int> | <chr> | <chr> | <fct> | <dbl> | <int> | |
| 1 | 2 | m | 23 | yes | Pre | Trained | 1195.40 | 20 |
| 2 | 3 | m | 26 | yes | Pre | Trained | 1681.00 | 30 |
| 3 | 5 | f | 20 | yes | Pre | Trained | 1427.20 | 0 |
| 4 | 6 | f | 21 | yes | Pre | Trained | 1798.30 | 5 |
| 5 | 7 | m | 27 | yes | Pre | Trained | 904.36 | 0 |
| 6 | 8 | m | 40 | yes | Pre | Trained | 504.40 | 20 |
Specify parameters¶
# ----------------------------------------------------------------------
# Specify items and parameters
# ----------------------------------------------------------------------
# From the dataset, specify subject ID, Dependent Variable, and 2 within-subject Factors
columns <- list(
sID = "sID",
DV = "ET",
Fc1 = "Type",
Fc2 = "Time"
)
# Define plot label names, tilte, colors and what type of error bars to show on the plot
param <- list(
y.label = "Execution Time (ms)",
Fc1.label = "Type",
Fc2.label = "Time",
cat.color = c('#df4a56', '#0272c6'),
errorbar = "ci" # can be either sd, se, or ci
)
# the plot title
param$title <- sprintf("%s x %s interaction", param$Fc2.label, param$Fc1.label)
Run the function¶
results <- rm_2by2_anova(dataOL, columns, param)
================================================================ CHECK ASSUMPTIONS ================================================================ Outliers # A tibble: 7 x 6 Fc1 Fc2 sID meanDV is.outlier is.extreme <fct> <fct> <fct> <dbl> <lgl> <lgl> 1 Trained Post 19 2444. TRUE FALSE 2 Trained Pre 17 3676. TRUE FALSE 3 Untrained Post 8 463. TRUE FALSE 4 Untrained Post 17 2551. TRUE FALSE 5 Untrained Post 19 2719. TRUE FALSE 6 Untrained Pre 17 3992 TRUE FALSE 7 Untrained Pre 19 3529. TRUE FALSE There are no extreme outliers. ----------------------------------------------- Normality # A tibble: 4 x 5 Fc1 Fc2 variable statistic p <fct> <fct> <chr> <dbl> <dbl> 1 Trained Post meanDV 0.957 0.602 2 Trained Pre meanDV 0.917 0.150 3 Untrained Post meanDV 0.919 0.164 4 Untrained Pre meanDV 0.902 0.0869 All levels are normally distributed. ================================================================ SUMMARY DESCRIPTIVES ================================================================ Fc1 Fc2 N meanDV sd se ci 1 Trained Post 16 1338.271 266.3100 66.57749 141.9066 2 Trained Pre 16 1865.416 265.2562 66.31405 141.3450 3 Untrained Post 16 1463.746 213.3426 53.33564 113.6822 4 Untrained Pre 16 1900.056 328.3329 82.08324 174.9563 ================================================================ 2x2 INTERACTION RESULTS ================================================================ Error: sID Df Sum Sq Mean Sq F value Pr(>F) Residuals 15 28566155 1904410 Error: sID:Fc1 Df Sum Sq Mean Sq F value Pr(>F) Fc1 1 102547 102547 1.864 0.192 Residuals 15 825140 55009 Error: sID:Fc2 Df Sum Sq Mean Sq F value Pr(>F) Fc2 1 3712982 3712982 25.17 0.000153 *** Residuals 15 2212669 147511 --- Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1 Error: sID:Fc1:Fc2 Df Sum Sq Mean Sq F value Pr(>F) Fc1:Fc2 1 33005 33005 1.791 0.201 Residuals 15 276434 18429 No significant Time x Type interaction: F(1,15) = 1.79, p = 0.2007469 ================================================================ Main Effects ================================================================ Main effect of Type: F(1,15) = 1.86, p = 0.1922710 ----------------------------------------------- Main effect of Time: F(1,15) = 25.17, p = 0.0001532 ================================================================ Post-hoc ================================================================ Pairwise comparisons grouped by Factor 1 # A tibble: 2 x 11 Fc1 .y. group1 group2 n1 n2 statistic df p p.adj * <fct> <chr> <chr> <chr> <int> <int> <dbl> <dbl> <dbl> <dbl> 1 Trained meanDV Post Pre 16 16 -5.32 15 0.0000852 0.0000852 2 Untrained meanDV Post Pre 16 16 -4.17 15 0.000817 0.000817 # ... with 1 more variable: p.adj.signif <chr> ----------------------------------------------- Pairwise comparisons grouped by Factor 2 # A tibble: 2 x 11 Fc2 .y. group1 group2 n1 n2 statistic df p p.adj p.adj.signif * <fct> <chr> <chr> <chr> <int> <int> <dbl> <dbl> <dbl> <dbl> <chr> 1 Post mean~ Train~ Untra~ 16 16 -3.05 15 0.008 0.008 ** 2 Pre mean~ Train~ Untra~ 16 16 -0.400 15 0.694 0.694 ns
Assumption check plots¶
options(repr.plot.width = 10, repr.plot.height = 5, repr.plot.res = 200) # change plot size
results$plot.assumption.checks
Results plot (with extras)¶
options(repr.plot.width = 7, repr.plot.height = 6, repr.plot.res = 200) # change plot size
# Adding post-hoc comparison stars
# Prepare the values
# post-hoc pairwise comparisons for each Fc1 level between the Fc2 levels
p.Fc1.L1 <-
ifelse(results$pwc1$p[1] < 0.05, stars.pval(results$pwc1$p[1]), "n.s.")
p.Fc1.L2 <-
ifelse(results$pwc1$p[2] < 0.05, stars.pval(results$pwc1$p[2]), "n.s.")
results$plot.anova +
# Add post-hoc pairwise comparisons
# might need to adjust the x and y positions
annotate(
"text",
x = 1.5,
y = 1400,
label = p.Fc1.L1,
color = param$cat.color[1],
size = 8
) +
annotate(
"text",
x = 1.5,
y = 1800,
label = p.Fc1.L2,
color = param$cat.color[2],
size = 8
) +
# add individual datapoints
geom_point(
colour = "black",
alpha = .3,
aes(fill = Fc1),
size = 2,
stroke = 0.5,
shape = 21,
position = position_jitterdodge(),
show.legend = FALSE
) +
# Show Pre first (instead of default alfabetical order)
scale_x_discrete(limits = c("Pre", "Post"))
Control what to display¶
By default, the function outputs a lot of results. If you don't want do display the output, you can 'hush' (suppress) it with a 'hush' function.
The 'hush' function¶
# hush function
# I got it from here https://stackoverflow.com/questions/2723034/suppress-output-of-a-function
hush <- function(code){
sink("NUL") # use /dev/null in UNIX
tmp = code
sink()
return(tmp)
}
Run the function without any output¶
results <- hush(rm_2by2_anova(dataOL, columns, param))
Choose what to output¶
Below is the list of possible output elements.
# results$outliers # results of identify_outliers function
# results$normality # results of shapiro_test
# results$plot.assumption.checks # boxplots and QQ plots in one plot
# results$dataSummary # Within-Subject descriptive summary of all 4 levels
# results$res.anova # ANOVA results of aov function
# results$one.way1 # Factor 1 Main effect results of anova_test function
# results$one.way2 # Factor 2 Main effect results of anova_test function
# results$pwc1 # Pairwise comparisons grouped by Factor 1
# results$pwc2 # Pairwise comparisons grouped by Factor 2
# results$plot.anova # the results plot
# for example, output ANOVA summary results
summary(results$res.anova)
Error: sID
Df Sum Sq Mean Sq F value Pr(>F)
Residuals 15 28566155 1904410
Error: sID:Fc1
Df Sum Sq Mean Sq F value Pr(>F)
Fc1 1 102547 102547 1.864 0.192
Residuals 15 825140 55009
Error: sID:Fc2
Df Sum Sq Mean Sq F value Pr(>F)
Fc2 1 3712982 3712982 25.17 0.000153 ***
Residuals 15 2212669 147511
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Error: sID:Fc1:Fc2
Df Sum Sq Mean Sq F value Pr(>F)
Fc1:Fc2 1 33005 33005 1.791 0.201
Residuals 15 276434 18429